3. Module 3¶
Module 3: Transcription Part II: What happens to the initial (pre-mRNA) transcript made by RNA pol II?
| Authors: | S. Catherine Silver Key (North Carolina Central University)
and Chiyedza Small (Medgar Evers College CUNY) |
|---|---|
| Last Update: | Sep 09, 2019 |
| Version: | 0.0.1 |
3.1. Introduction¶
In Module 2, you identified the transcription start site (TSS) for the A isoform of the tra gene (tra-RA). In this module, we will explore each of the three steps of pre-mRNA processing.
3.1.1. Setting up our Browser page (review):¶
- Open a new web browser window and go to the UCSC Genome Browser Mirror
site at http://gander.wustl.edu/. Follow the instructions given in
Module 1 to navigate to the
contig1project in the D. melanogasterJuly 2014 (Gene)assembly. - As you may remember from Module 1,
contig1 is derived from chr3L in the D. melanogaster genome. This
contig contains three different genes (CG32165, spd-2, and
tra). Enter
contig1:9,500-11,000into the “enter position or search terms” textbox and then click on thegobutton to navigate to the genomic region surrounding the tra gene. - Because the Genome Browser remembers your previous display settings,
you should click on the
default tracksbutton to reset the display to the default settings. Change the display mode for the “Base Position” track tofulland verify that the “FlyBase Genes” track is set topack. Click on therefreshbutton. - Scroll down to the “RNA Seq Tracks” section and then click on the
RNA-Seq Coveragelink. Change the track display settings to the following, as we did in Module 2:- Change the “Display mode” field to
full - Select the “Data view scaling” field to
use vertical viewing range setting - Change the “max” field under “Vertical viewing range” to
37 - Under the “List subtracks” section, select BOTH the
Adult Femalesand theAdult Malessubtracks (Select the check box next to subtrack to turn the subtrack on.) - Click on the
Submitbutton. Verify that the RNA-Seq Coverage track on the browser page is set tofull.
- Change the “Display mode” field to
3.2. Investigation: mRNA processing¶
Learning Objectives
- Use the genome browser to identify the relationships among; pre-mRNA, 5’ capping, 3’ polyadenylation, splicing, mRNA.
- Describe how the transcript generated by RNA polymerase II (the pre-mRNA) is processed to become mature mRNA, using the sequence signals identified in Module 2.
The processing of pre-mRNA into mRNA involves three key steps (Figure 3.1)
- The addition of a 5’ cap
- The addition of a 3’ poly(A) tail
- The removal of introns through splicing
Removal of the introns during this process results in adjacent exons being brought together in the final mRNA message.
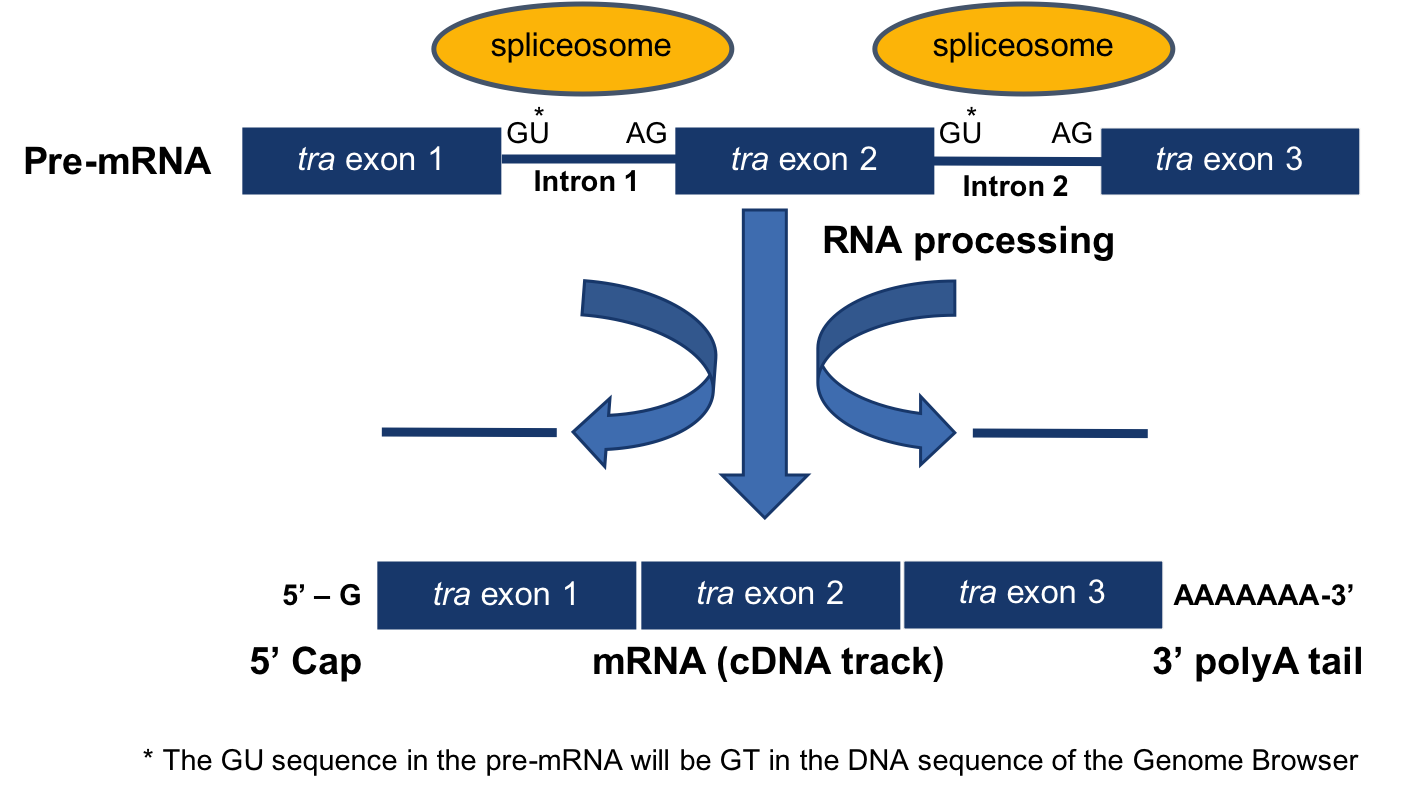
Figure 3.1. Diagram of mRNA processing that converts a pre-mRNA to a processed mRNA.
3.2.1. Addition of a 5’ cap¶
The first step in pre-mRNA processing occurs at the 5’ end
of a messenger RNA. Recall that mRNA is synthesized in a 5’ to 3’
direction, so the 5’ end of the mRNA was synthesized first. Let’s
examine the beginning of the tra gene. Type contig1:9,825-9,870
into the “enter position or search terms” textbox and then click on
the go button.
In Module 2, we identified the transcription start site (TSS) of the A isoform of tra at position 9,851.
To show the TSS’s that have been annotated by the modENCODE project,
scroll down to the “Genes and Gene Prediction Tracks” and change the
display mode for the “TSS Annotations” track to pack, and click
refresh. The modENCODE project looked for TSSs by using a chemical
method to tag the special structure that occurs at 5’ ends of
transcript, fishing out the RNA molecules that carried these tags, and
mapping the sequence back to the genome, a method called “CAGE” (cap
analysis of gene expression).
In addition, we will also display the “D. mel. cDNAs” track (also
under the “Genes and Gene Prediction Tracks” section); change this to
pack. This track shows the alignment of D. melanogaster
cDNAs (complementary DNAs, made by copying the mRNA)
that have been sequenced by the Berkeley Drosophila Genome Project
(BDGP). Click on the refresh button
(Figure 3.2). These two tracks both
provide an analysis based on the RNA population, and mapping the
positions of these sequences indicates where the transcript started.
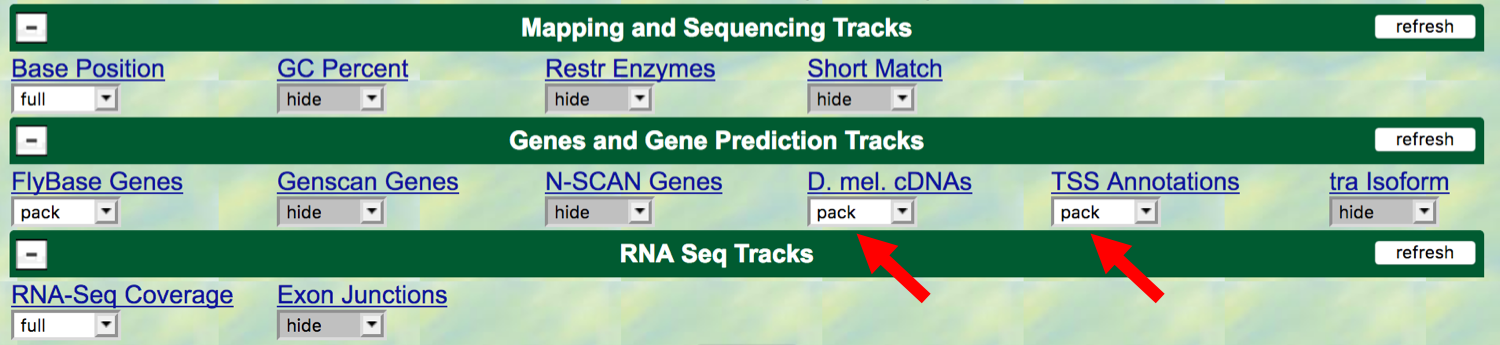
Figure 3.2. Change the display modes of the “D. mel. cDNAs” and “TSS Annotations” tracks to “pack”.
Remember from Module 2 that we also found a match to TCAKTY, a common initiation signal just upstream, at 9,834 (display this using the “Short Match” track). All of these pieces of evidence argue for a TSS in this region.
The new Genome Browser image (Figure 3.3) shows the 5’ end of the pre-mRNA transcript (i.e. the start of transcription) based on the CAGE experiment (modENCODE track) with the additional lines of support. On this end of the pre-mRNA, a modified guanine nucleotide (7mG) is added to the nucleotide at position 9,851, forming the 5’ cap. Note that this additional nucleotide is NOT visible in the DNA track. It is added AFTER the transcript is made. This is the first step in pre-mRNA processing: capping.
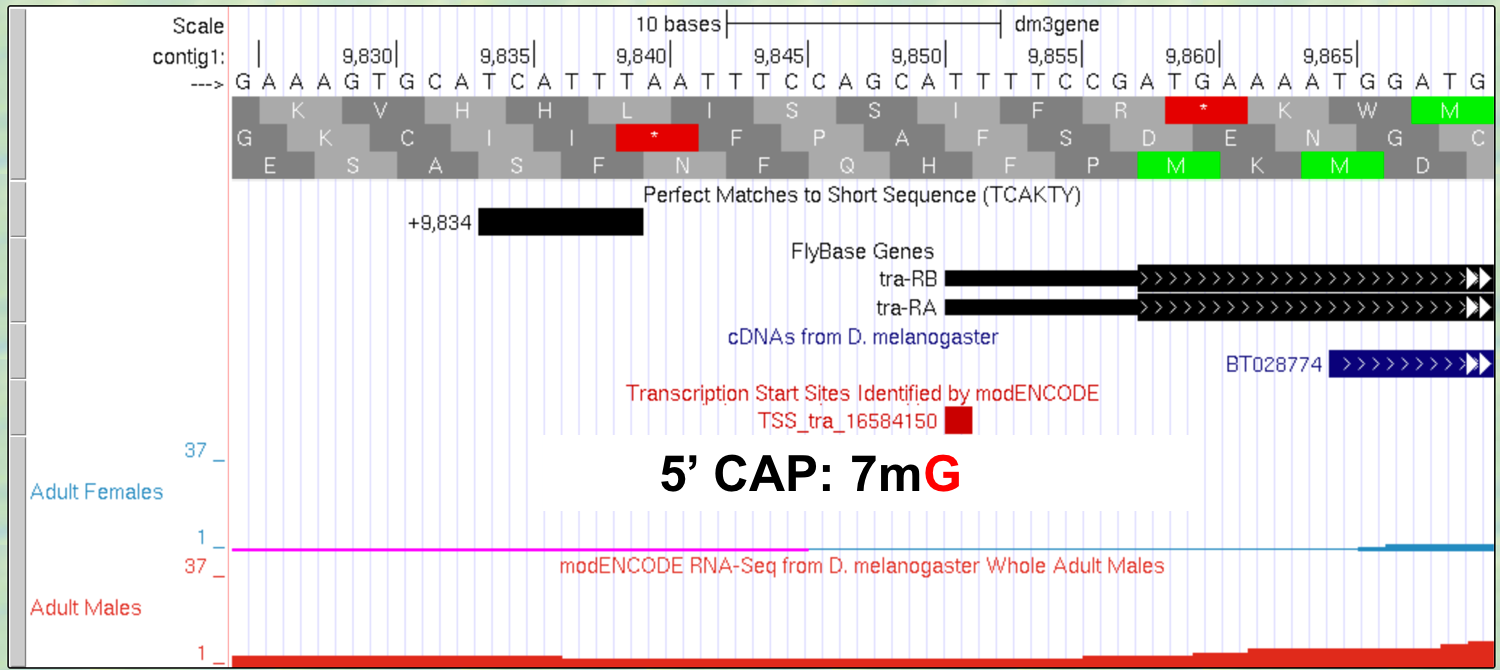
Figure 3.3. Addition of a 5’ cap to the 5’ end of the transcript.
Question 1
What is the coordinate of the first nucleotide that is transcribed? In the DNA sequence, is it an A, C, T or G?
Question 2
What are the coordinates for the start codon that codes for the first amino acid of the A isoform of the tra gene? (Assume reading frame +3.)
Question 3
The region of the transcript from the 5’ cap to the nucleotide just upstream of the start codon is called the 5’ untranslated region (5’UTR) because it is part of the transcript that is not translated. How long (in ribonucleotides) is the 5’UTR?
3.2.2. Addition of a 3’ poly(A) tail¶
The second step in pre-mRNA processing is polyadenylation.
- To view the 3’ end of the tra-RA gene, change the
“enter position or search terms” field to
contig1:10,633-11,000and then click on thegobutton.
Polyadenylation means that many (poly) adenine nucleotides (ribonucleotides) are added to the 3’ end of the pre-mRNA AFTER transcription termination. This generates a poly-A tail (typically ~20 to ~250 As) that will be retained in the final mRNA but it is not present in the “Base Position” track of the Genome Browser. This is because the poly-A tail does not exist in the DNA template but is simply added to the RNA by a special polymerase as a long run of adenine nucleotides.
Our previous analysis in Module 1 has shown that the last coding exon of tra-RA is in frame +2 and the stop codon is located at 10,754-10,756. We can use the Genome Browser to determine the end of the tra-RA transcript indicated by the cDNA track (in blue). (Note that this aligns with the cDNA although there is some discrepancy between the two as to the exact end of the transcript.)
Question 4
How long (in base pairs) is this 3’ untranslated region (3’UTR) as indicated by the cDNA track (in blue)?
Question 5
Zoom into the 3’ end of the FlyBase Gene, near the termination site. What is the longest stretch of A nucleotides that you observe?
Question 6
Do your findings support the conclusion that the poly(A) sequence observed in the mature mRNA transcript is not in the template DNA?
- Perform a “Short Match” search for the poly-A signal using the protocol you learned in Module 2. This search should place the poly-A signal at 10,818-10,823 (Figure 3.4). As mentioned in Module 2, the transcript is cleaved 11 to 30 nucleotides downstream of the poly(A) signal sequence, and then 150-200 adenines are added to the pre-mRNA. The nucleotides between the stop codon and the end of the poly-A tail comprise the 3’ UTR.
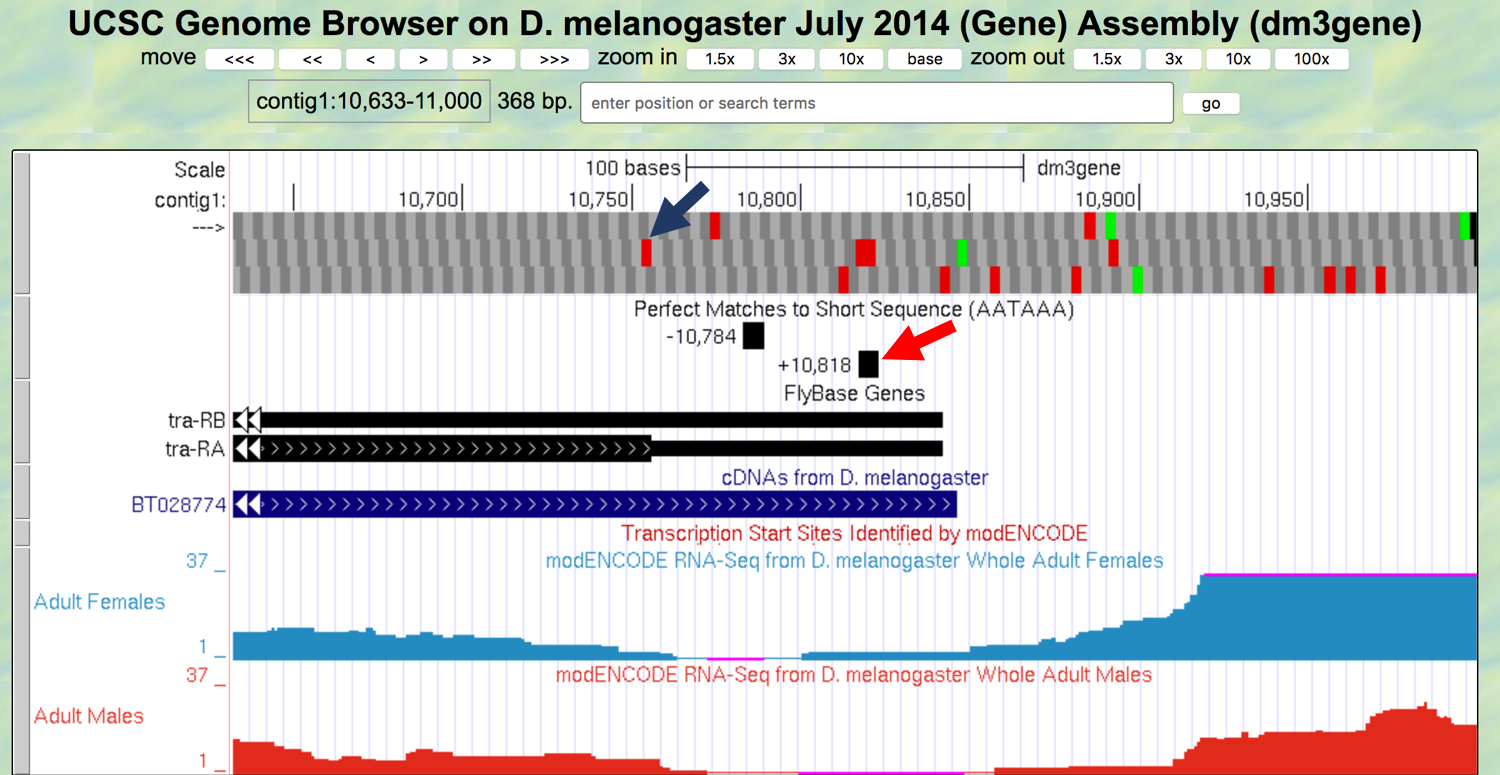
Figure 3.4. Previous analysis placed the stop codon for the A isoform of tra in frame +2 (blue arrow) and the poly-A signal at 10,818-10,823 (red arrow).
We can see the polyadenylation sequence that is associated with the processed mRNA by examining the cDNA (BT028774) that has been aligned to this region.
- Click on the
BT028774feature under the “cDNAs from D. melanogaster” track and then click on theView details of parts of alignment within browser windowlink (Figure 3.5).
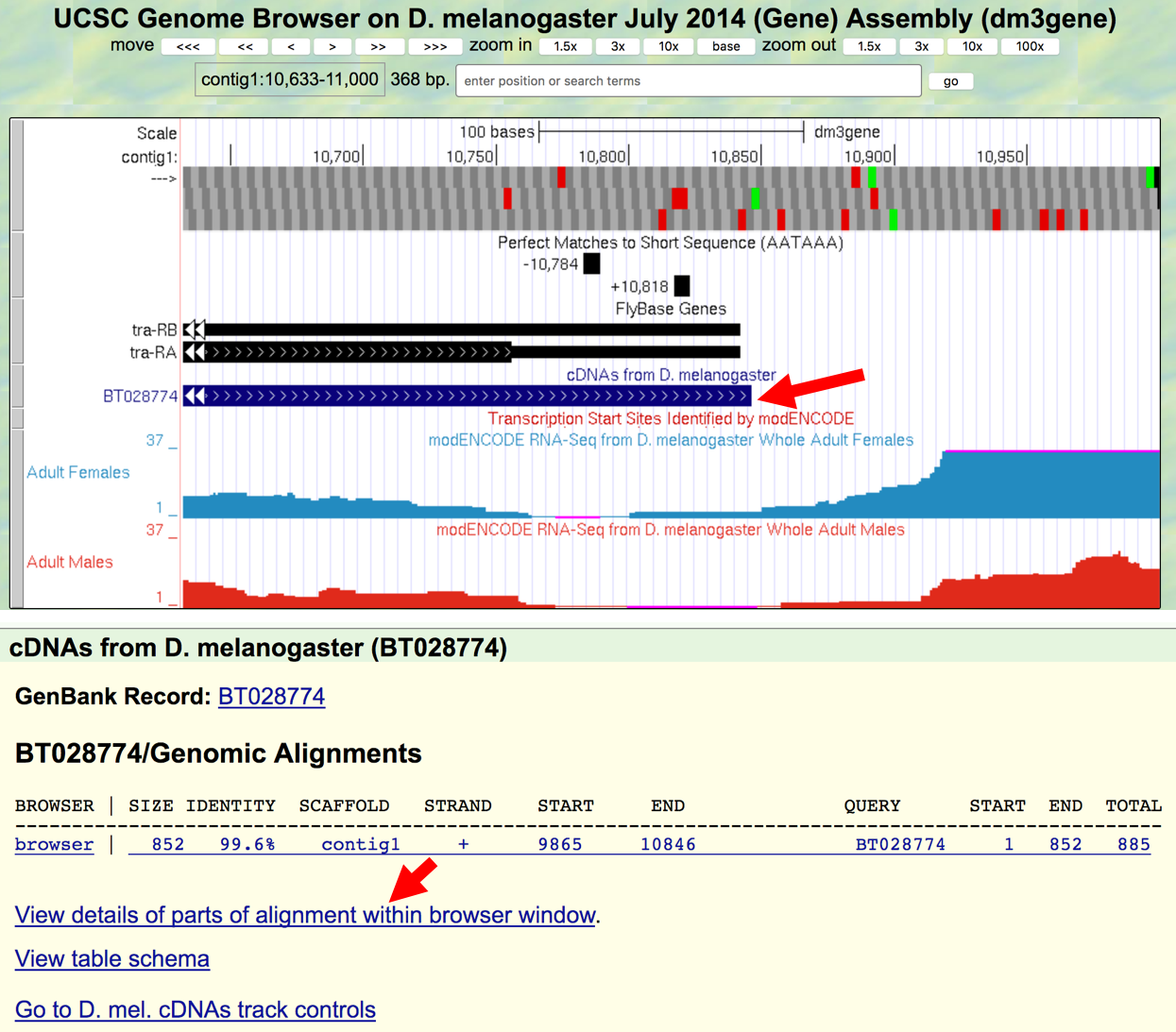
Figure 3.5. Examine the alignment of D. melanogaster cDNA BT028774 against contig1.
The next figure shows the actual alignment between the D. melanogaster cDNA BT028774 and the genomic sequence in contig1 (Figure 3.6). Nucleotides that are identical between the two sequences are shown in blue capital letters while nucleotides that differ are shown as black lowercase letters. The light blue bases denote the start and the end of the gap in the alignment. The side-by-side alignment shows the pairwise alignment between the cDNA (top) and the contig1 sequence (bottom) within the viewing region (i.e. contig1:10,633-11,000).

Figure 3.6. Alignment of the D. melanogaster cDNA BT028774 with the end of contig1.
Question 7
Scroll up to the cDNA BT028774 area. After which coordinate
(number in the cDNA) do you see the polyadenylation track (in lower
case black letters)?
Question 8
How many “A” ribonucleotides have been added to the tra mRNA (represented in the cDNA)?
Question 9
Locate the AATAAA termination signal in the cDNA sequence. How
many nucleotides 3’ of the final “A” in the signal sequence does the
poly(A) run start? (This number should be between 11–30 nucleotides.)
3.2.3. Removal of introns through splicing¶
The final step in pre-mRNA processing is splicing out of introns and merging adjacent exons into one continuous open reading frame so that the mRNA is ready for translation into a protein.
- Change the “enter position or search terms” field to
contig1:9,870-10,170and then click on thegobutton to navigate to the first intron of the tra-RA transcription (Figure 3.7).
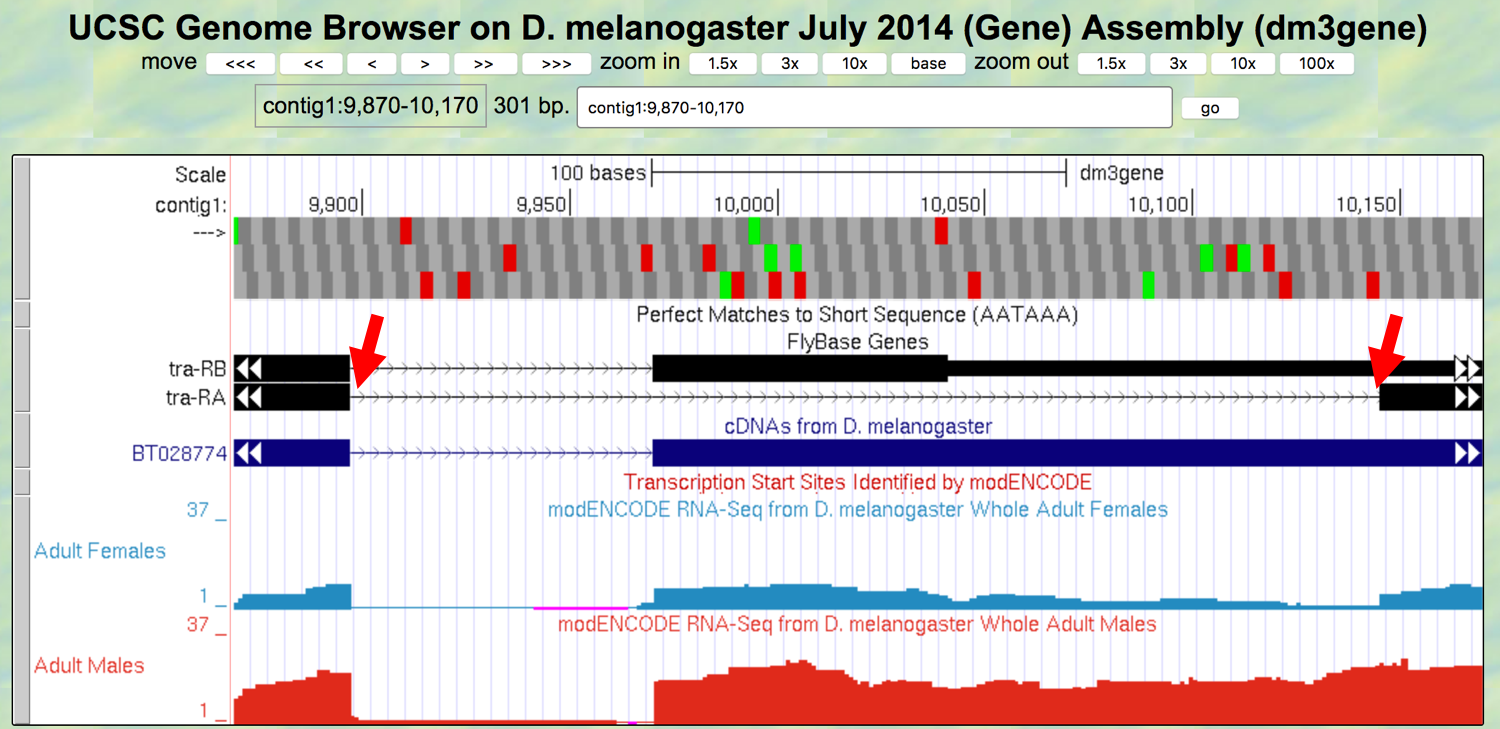
Figure 3.7. The genomic region surrounding the first intron (red arrows) of tra-RA.
- Zoom in to the region near the end of the first exon of tra-RA.
Question 10
Which two nucleotides are found just after the end of the first exon of tra-RA? Repeat this determination, identifying the two nucleotides at the start of intron 2 of tra-RA.
These two nucleotides are a signal for splicing to occur at the 5’ end of an intron; these represent the first two bases of the intron, often called the donor site (or 5’ splice site).
Question 11
At which base does exon 1 end?
- Zoom out and then zoom in to the region near the beginning of the second exon of tra-RA.
Question 12
Which two nucleotides are found right before the start of tra-RA exon 2?
- Zoom out and then zoom in to the region near the beginning of the third exon of tra-RA.
Question 13
Which two nucleotides are found right before the start of tra-RA exon 3?
These two nucleotides are the signal for splicing out of the 3’ end of the intron, often called the acceptor site (or 3’ splice site). These represent the last two bases of the intron.
Question 14
At which base does exon 2 of tra-RA begin? What is its coordinate?
3.3. Summary¶
In this module, we learned about the three key steps that are involved in converting the pre-mRNA into a mature mRNA.
- The addition of a 5’ cap
- The addition of a 3’ poly(A) tail
- The removal of introns through splicing
Note
Introns are removed during this process and adjacent exons are brought together in the mRNA message.
After mRNA processing, the mature mRNA (tra-RA) can now exit the nucleus so that it can be translated into a protein (tra-PA) by the cytoplasmic ribosomes.
Summary Question 1
Describe how the transcript generated by RNA polymerase II (the pre-mRNA) is processed to become mature mRNA, using the sequence signals identified in Module 2.